Parameter Setting for Pathway Analysis
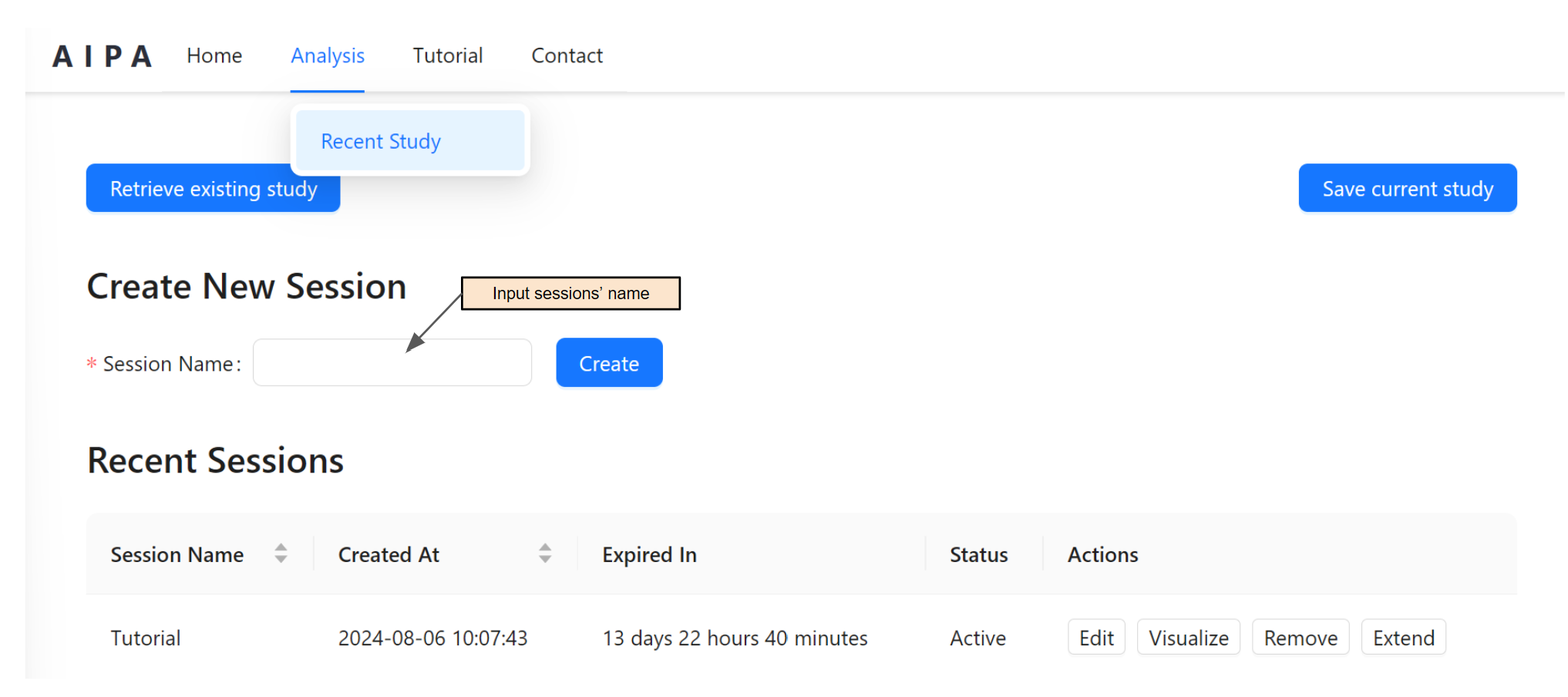
All analyses are performed on the Analysis page. To access the Analysis page, either click Begin Your Analysis on the homepage or select Recent Study under the Analysis menu. In this page, users are able to create new sessions, switch or delete sessions, visualize analysis session's results, or visualize local data.
To create a new session, input a session name and press create.
Once a session is created, users can edit the analysis' name (Step 1). Next, users select input type for the analysis in Step 2. One session can analyze multiple datasets by adding multiple analyses (Step 3 & 4). This is especially helpful when users want to analyze multiple datasets at the same time for meta-analysis.
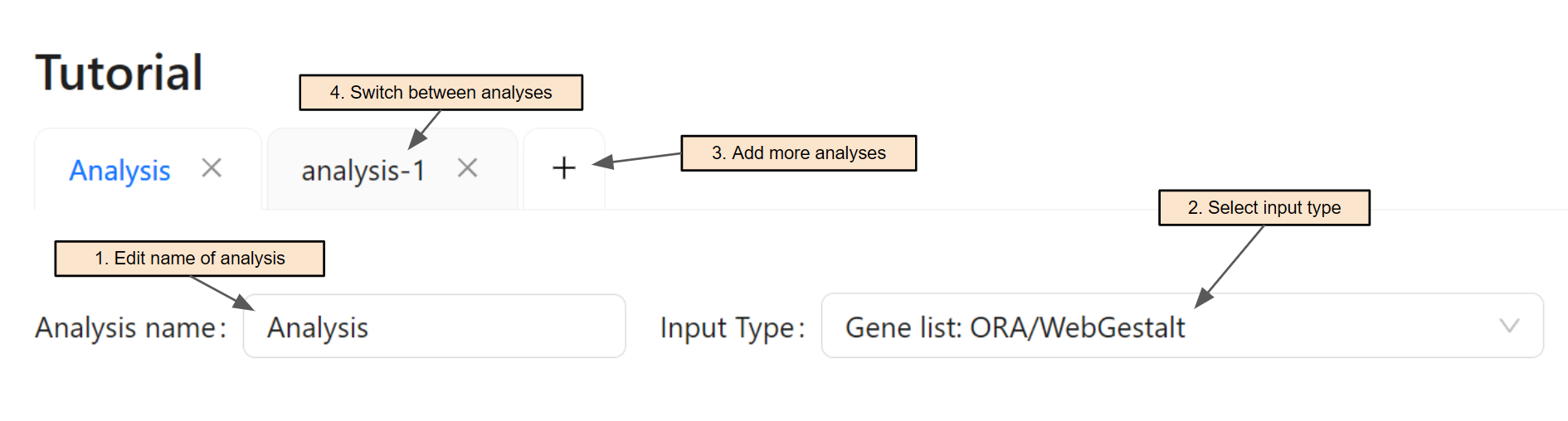
For each input type, see the corresponding sections below for its tutorial.
Gene list
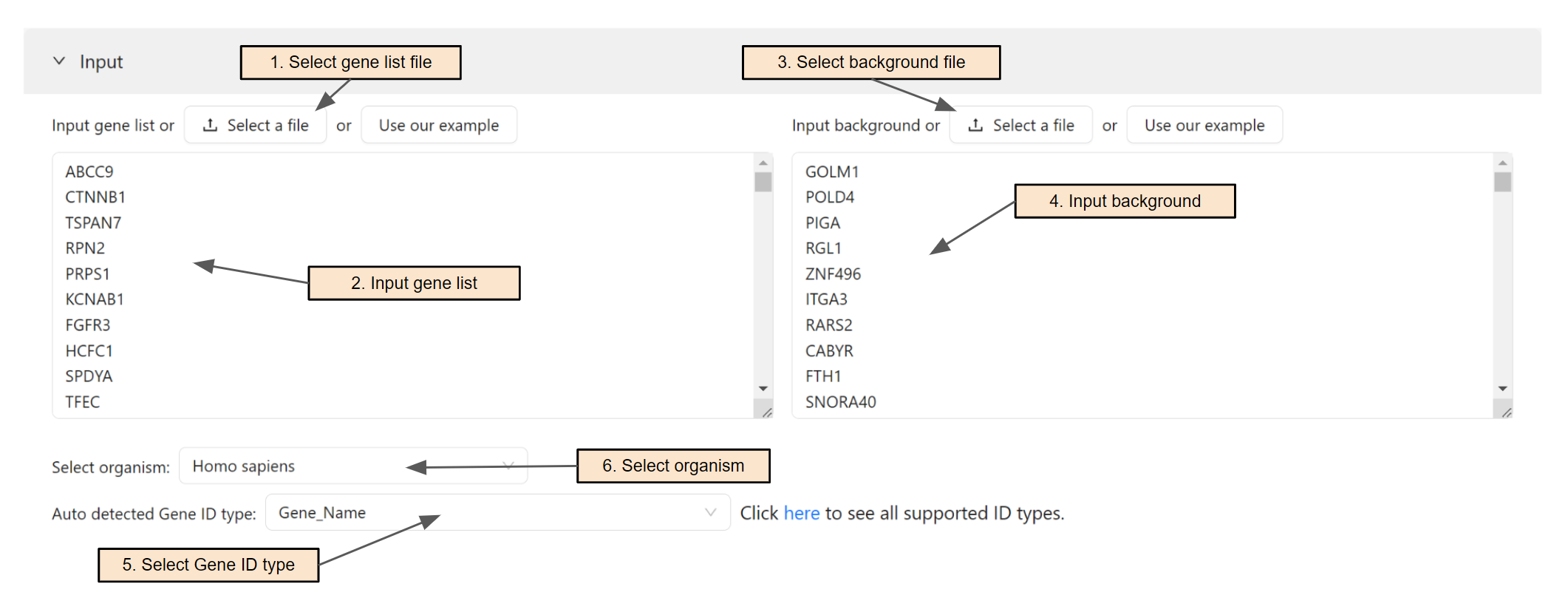
Input gene list and background
Users can select a gene list file (Step 1) or paste the gene list directly into the text area (Step 2). Users can also select a background file (Step 3) or paste the background directly into the text area (Step 4). If a file is selected, it's content will be automatically parsed into the text area.
The system will automatically detect the input ID type. Users can change the ID type of the input in Step 5.
Users will then select the organism for the analysis. To search for an organism, simply enter text in the search box (Step 6). To see all supported organisms, please visit the Data source page.
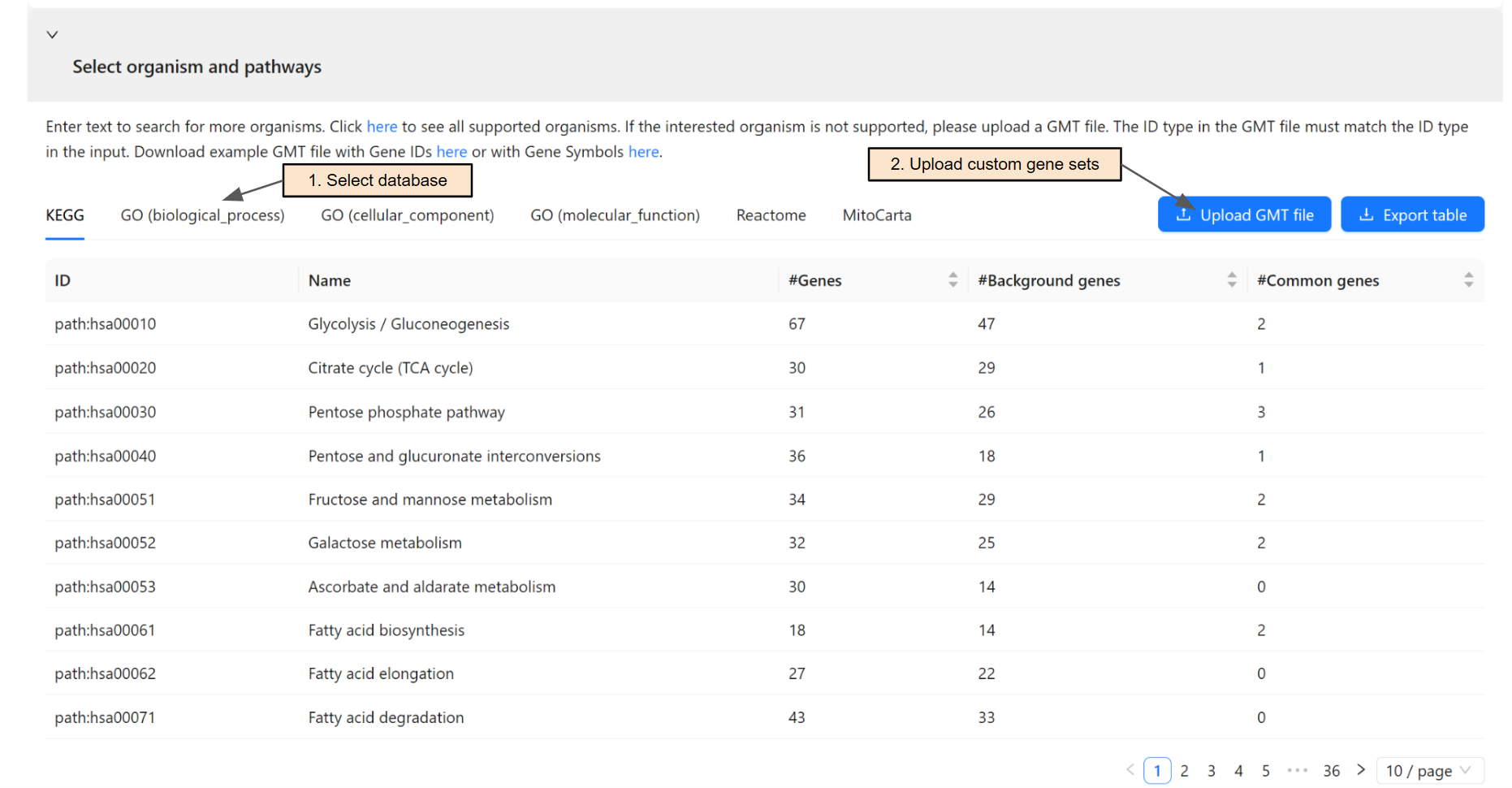
Select pathways and gene sets for the gene list analysis
Once an organism is selected, the system will retrieve gene sets from KEGG database, Gene Ontology, Reactome, and MitoCarta. Users can switch among databases using the corresponding tab (Step 1).
Users can upload a GMT file for custom gene sets (Step 2) if their database of interest is not supported. The ID type in the GMT file must match the ID type in the input.
Gene list and fold change
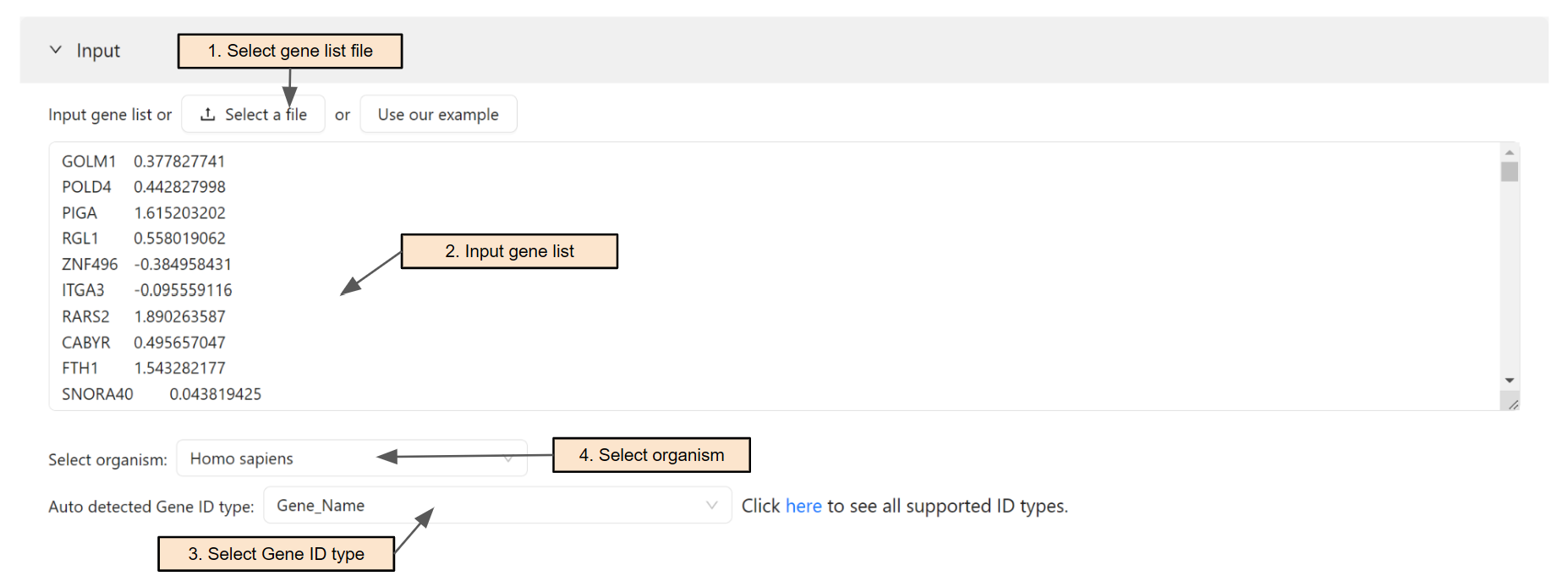
Input gene list and fold change
Users can select a gene list and fold change file (Step 1) or paste the gene list and their fold change directly into the text area (Step 2). If a file is selected, it's content will be automatically parsed into the text area.
The system will automatically detect the input ID type. Users can change the ID type of the input in Step 3.
Users will then select the organism for the analysis. To search for an organism, simply enter text in the search box (Step 4). To see all supported organisms, please visit the Data source page.
Select pathways and gene sets for the analysis using gene list and fold change
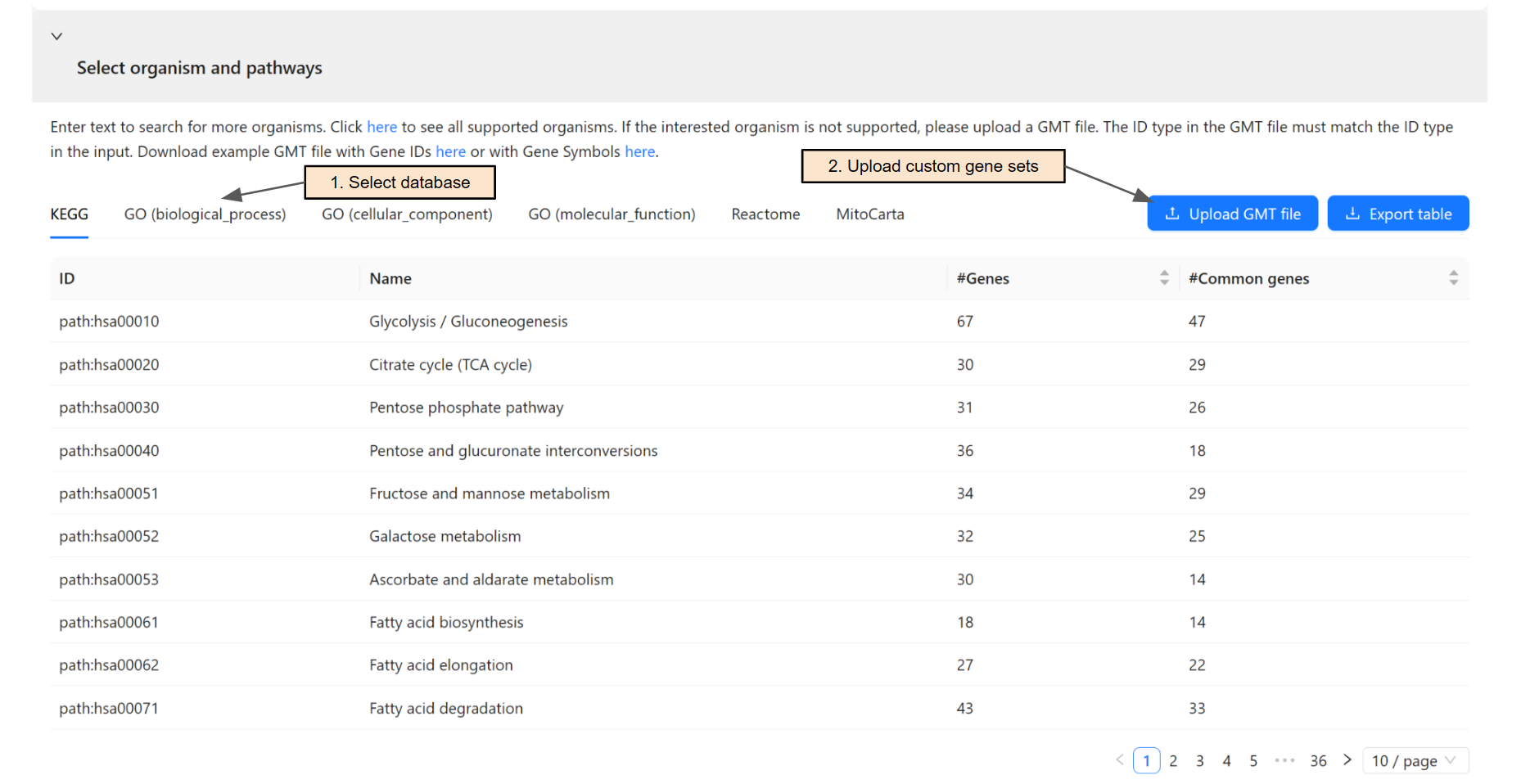
Once an organism is selected, the system will retrieve gene sets from KEGG database, Gene Ontology, Reactome, and MitoCarta. Users can switch among databases using the corresponding tab (Step 1).
Users can upload a GMT file for custom gene sets (Step 2) if their database of interest is not supported. The ID type in the GMT file must match the ID type in the input.
Select pathway analysis methods using gene list and fold change
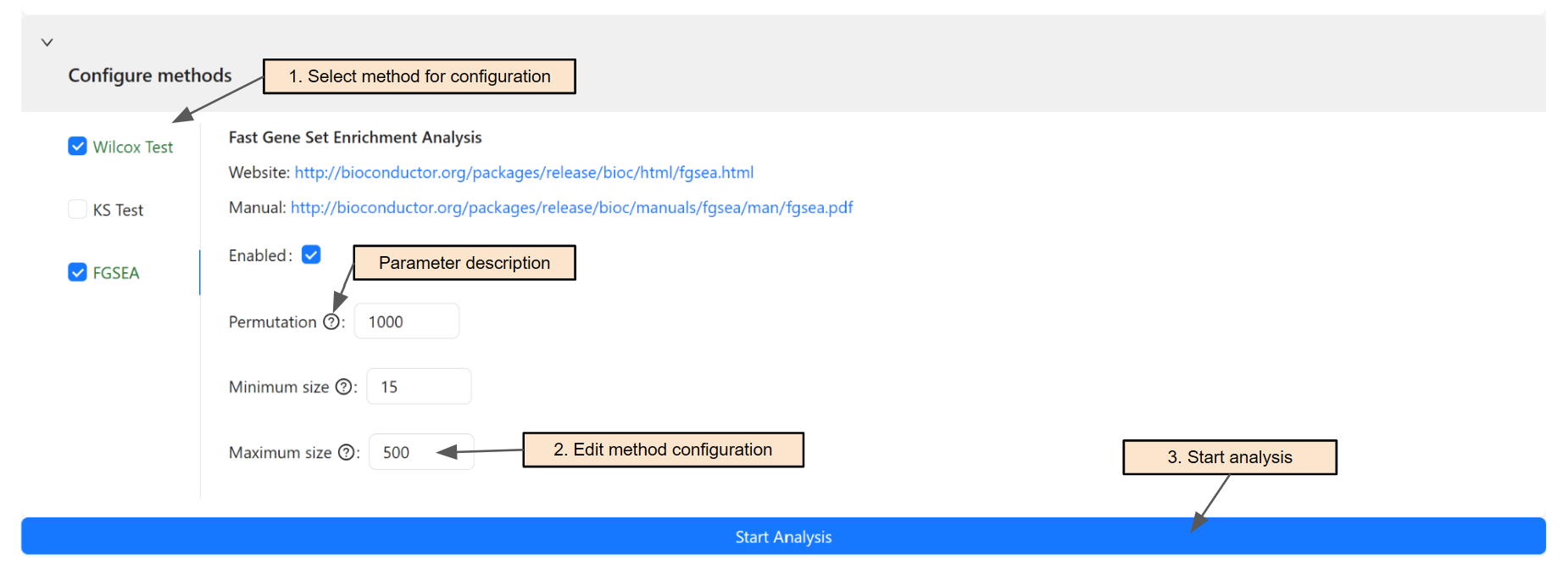
Users can analyze the input data using multiple methods. To select the method for analysis and change its configuration, select the corresponding tab (Step 1 & 2). Users can hover over the question mark next to the parameter name to view the description for that parameter.
Finally, click on Start analysis (Step 3) to start the pathway analysis using selected methods.
Note: After clicking the Start analysis button, the analysis will be computed for only the current selected dataset. Users need to repeat the steps above for each dataset included in the analysis.
Currently, supported methods are:
Wilcox Test: Wilcoxon signed-rank test
KS Test: Kolmogorov-Smirnov test
FGSEA: Fast Gene Set Enrichment Analysis
Website: http://bioconductor.org/packages/release/bioc/html/fgsea.html
Manual: http://bioconductor.org/packages/release/bioc/manuals/fgsea/man/fgsea.pdf
Expression data
Select data for the analysis
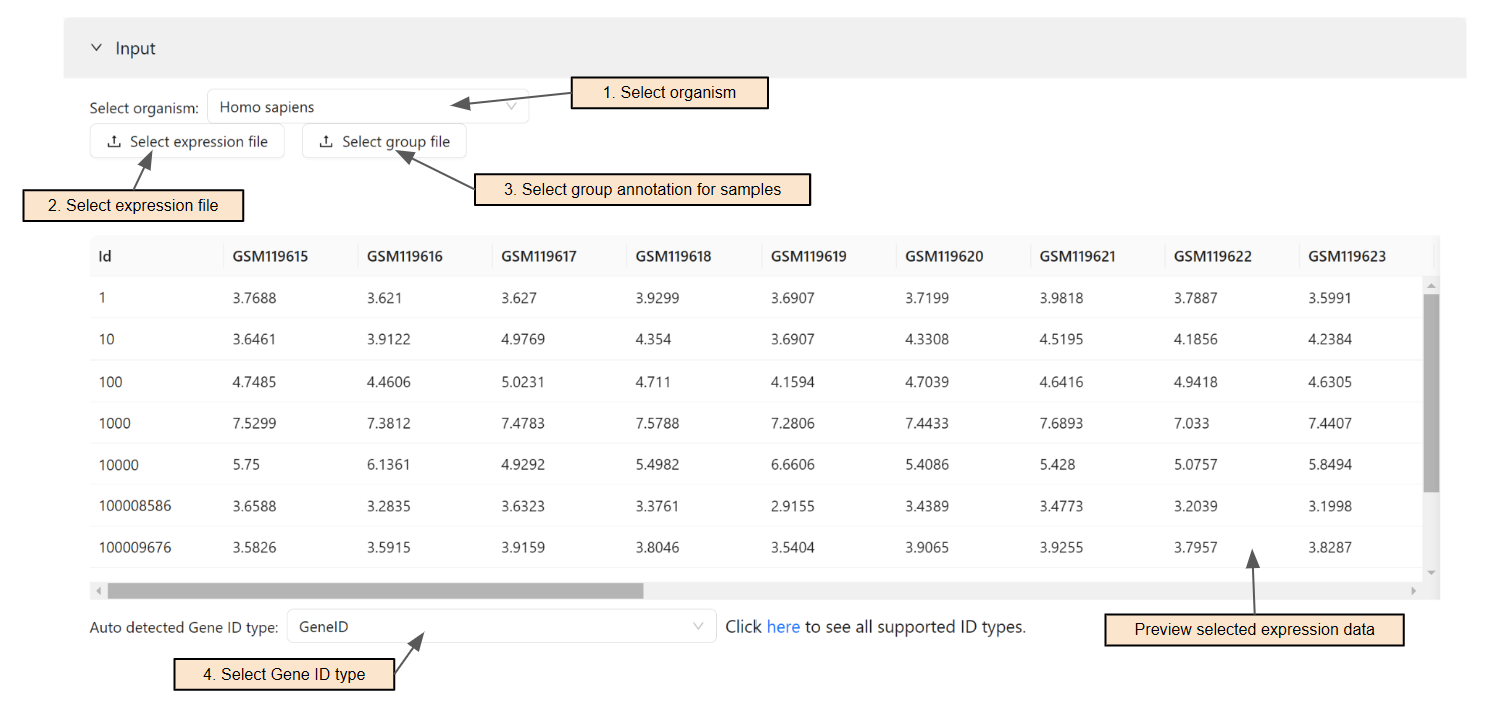
Users can select the organism for the analysis. To search for an organism, simply enter text in the search box (Step 1). To see all supported organisms, please visit the Data source page.
Users can select expression file (Step 2) and group file (Step 3) for the analysis. A popup similar to File manager will show up to select files. After that, a preview of the selected data will show up for users to verify the data
The system will automatically detect the input ID type. Users can change the ID type of the input in Step 4.
The system will automatically detect the input ID type. Users can change the ID type of the input in Step 3.
Users will then select the organism for the analysis. To search for an organism, simply enter text in the search box (Step 4). To see all supported organisms, please visit the Data source* page.
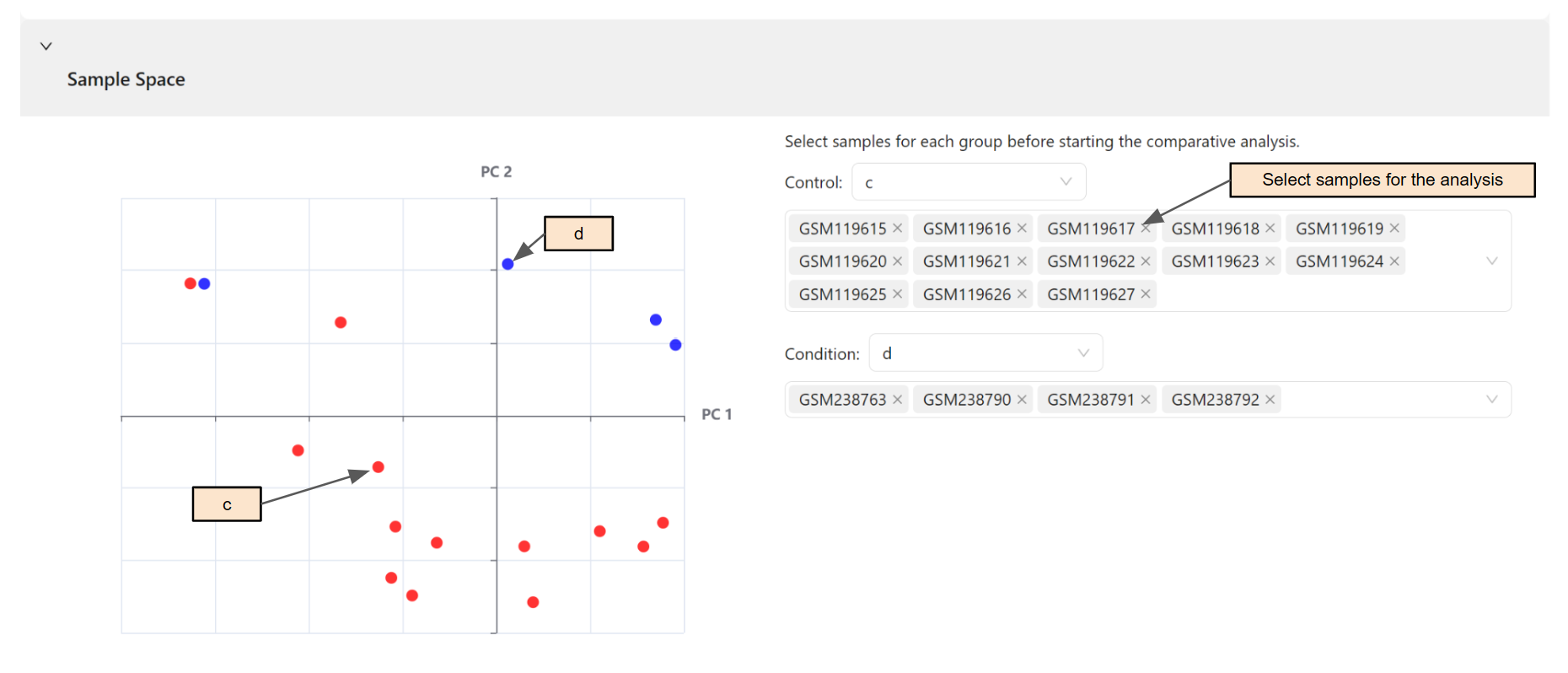
Select samples and genes for the analysis
Once expression data for the analysis is selected, a t-SNE plot will be presented for an overview of the data landscape.
Users then must choose samples for the analysis, divided into two groups: control(c) and disease(d).
If group data file is selected in the previous step, users can easily select all samples in a group by selecting the group in the selection box. Users can also remove samples from a group by manually moving the samples to the "Exclude box" to excluded them from the analysis.
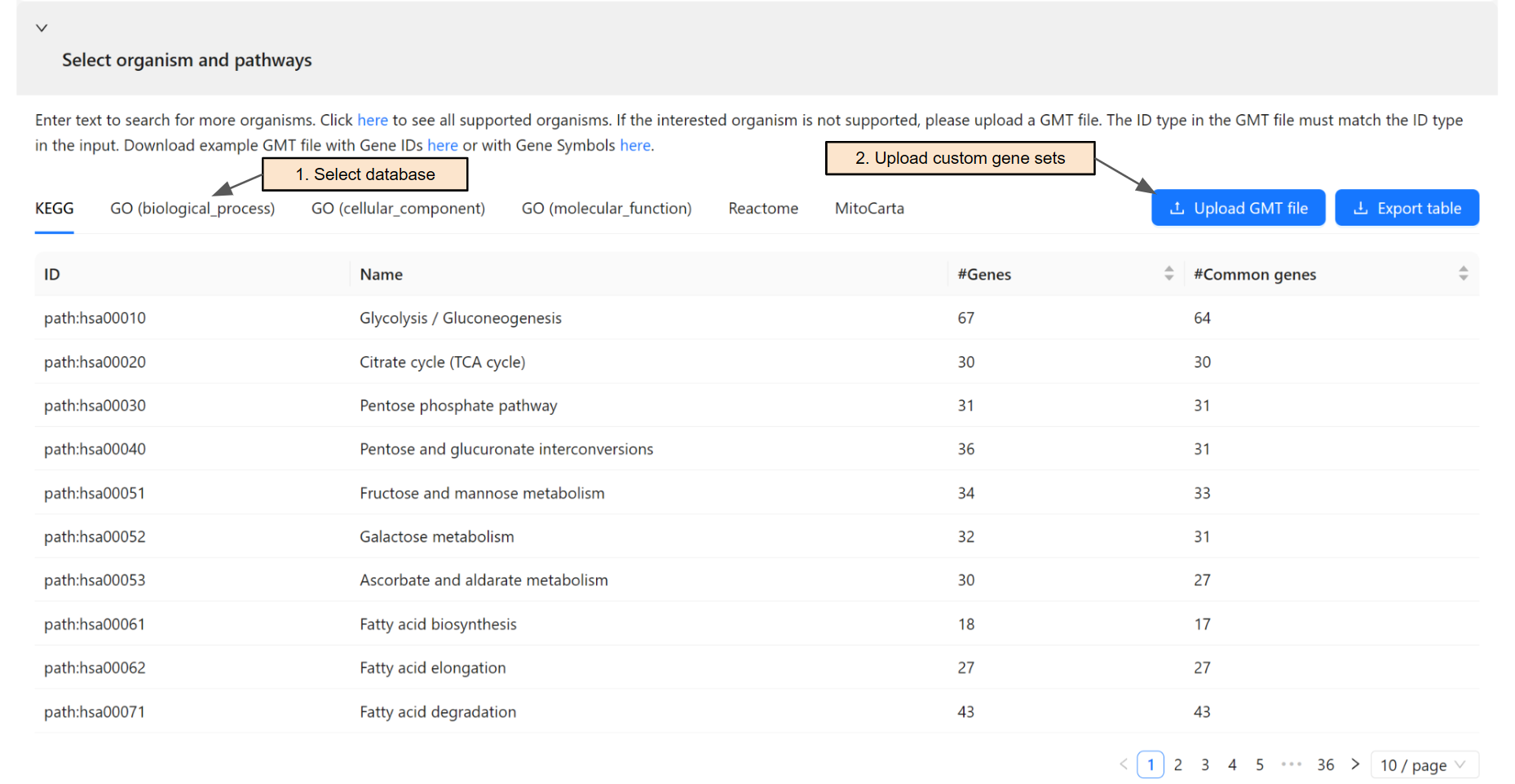
Select pathways and gene sets for the analysis
Once an organism is selected, the system will retrieve gene sets from KEGG database, Gene Ontology, Reactome, and MitoCarta. Users can switch among databases using the corresponding tab (Step 1).
Users can upload a GMT file for custom gene sets (Step 2) if their database of interest is not supported. The ID type in the GMT file must match the ID type in the input.
Select pathway analysis methods
Users can analyze the input data using multiple methods. To select the method for analysis and change its configuration, select the corresponding tab (Step 1 & 2). Users can hover over the question mark next to the parameter name to view the description for that parameter.
Finally, click on Start analysis (Step 3) to start the pathway analysis using selected methods.
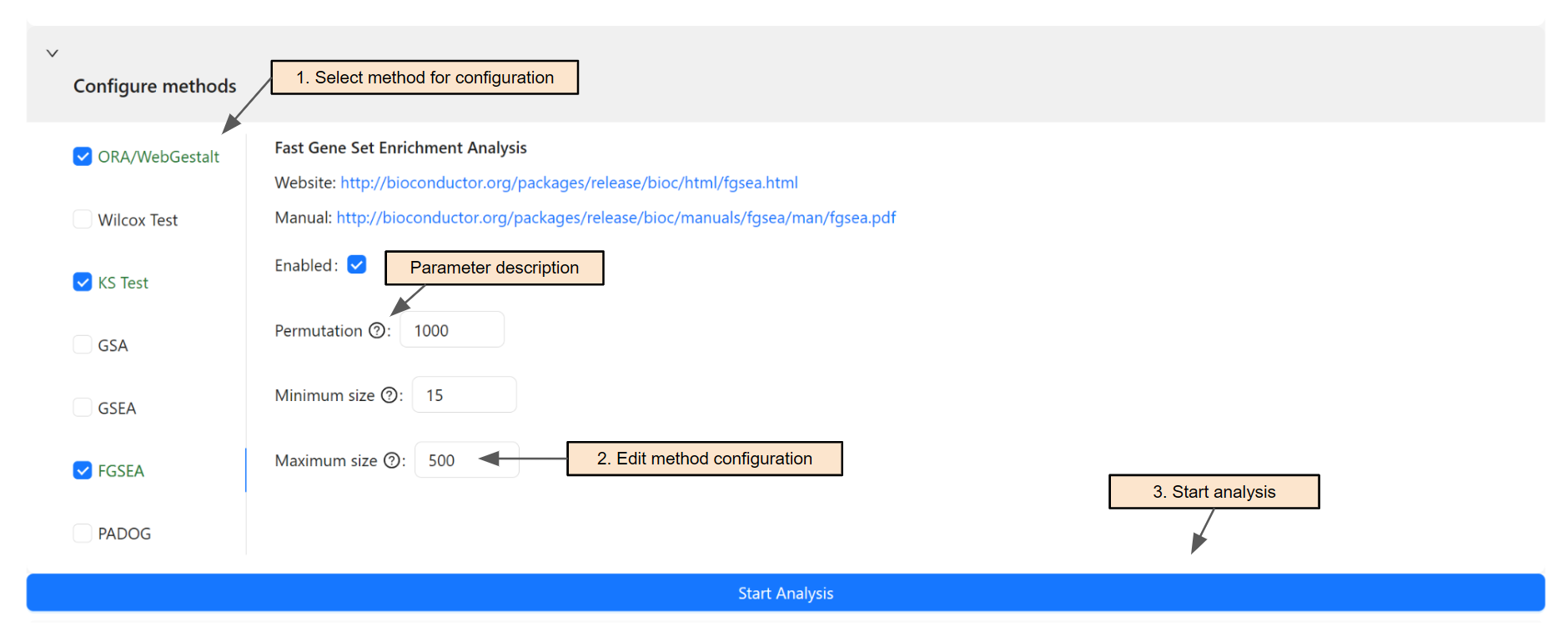
Note: After clicking the Start analysis button, the analysis will be computed for only the current selected dataset. Users will need to repeat the steps above for each dataset included in the analysis